Separating Nucleic Acids Using HPLC

Chromatography has been used as a separation and analysis tool since ca. 1903–6 Russia. Although it has had a diverse assortment of applications, it has only been used in molecular biology applications on rare occasions. Recently, Transgenomic Inc. (San Jose, CA; 408-514-3132) has developed a series of advanced HPLC systems. With these tools, chromatography may replace traditional electrophoresis methods in some bioresearch laboratories.
Contents
HPLC vs. Electrophoresis
How The Transgenomic System Works
Applications
HPLC vs. Electrophoresis (Back to Top)
Traditionally, bioresearchers have selected various permutations of electrophoretic separations over HPLC techniques. Despite this preference, several new factors are now making HPLC quite appealing to the bioresearch community:
- Automation. In bioresearch, the standard sample tube is based on the microtiter format. A microtiter format autosampler is standard for the Transgenomic system. The system loads and separates all samples without operator intervention.
- Minimal Set-up. Gel systems require scientists to prepare a gel and set up a system for each run. This has been somewhat obviated by precast minigels, but the use of such gels is limited by application. With HPLC, a pre-packed cartridge is installed that is suitable for 4000+ samples. Maintenance is achieved by modifying the buffer and adjusting the flow through the system.
- Precise control of separation conditions. In the Transgenomic system, parameters including flow rate, buffer concentration, and temperature are controlled by the computer through software that helps users to precisely predict parameters based upon application and, in some cases, fragment sequence.
- Sample retrieval. Upon completion of a gel run, samples are separated and suspended in the gel matrix. The gel becomes a sample contaminant requiring additional purification products and steps, resulting in a reduction in recovery. With HPLC, samples are separated and detected. They can then be retrieved in the downstream buffer flow. Elution buffer does not interfere with subsequent applications.
- Quantification. With nucleic acid chromatography, both UV and fluorescence detection are available. Both offer direct quantification of fragments in samples during separation.
How The Transgenomic System Works (Back to Top)
Transgenomic's HPLC systems are based on a proprietary separation technology called the DNASep column. These cartridges incorporate non-porous poly (styrene-divinylbenzene) copolymer beads to create a highly hydrophobic matrix as the stationary phase. When combined with an ion-pair reagent as the mobile phase (TEAA), a dynamic ion-change separation environment is created. This is called ion-pair reversed-phase HPLC (IP-RP-HPLC).

Transgenomic's Wave nucleic acid fragment
analysis system utilizes DNASep technology.
The principles of IP-RP-HPLC are quite simple. The column (or stationary phase) is hydrophobic, while nucleic acids are negatively charged at neutral pH. This means that nucleic acid will be repelled by the column surface. TEAA (the mobile phase) is a molecule with both a positively charged ammonium ion and charge-neutral alkyl groups. Thus the two different moieties of the TEAA can interact with the nucleic acid and the DNASep matrix, creating a chemical bridge. An organic solvent--acetonitrile in this case--interrupts the TEAA/DNASep complex of nucleic acid. By applying a precise gradient of increasing concentration of acetonitrile to the mobile phase, nucleic acids can be selectively eluted based upon how tightly they are bound, thus affecting a separation.
Either UV absorption or fluorescence, depending on the specific system, is used to detect eluted sample components. UV detection offers precise quantification, while the fluorescence allows for selective detection and improved sensitivity in addition to quantification. As an option, separated, purified fragments can be collected in an automated, chilled, microtiter format fragment collector,
Transgenomic's HPLC systems are protected by U.S. patent numbers 5,585,236 and 5,772,889.
Applications (Back to Top)
The temperature at which the separation occurs can be precisely controlled and results in three distinct modes of operation and a wide range of possible applications. The three operation modes are non-denaturing, partial denaturing, and full denaturing.
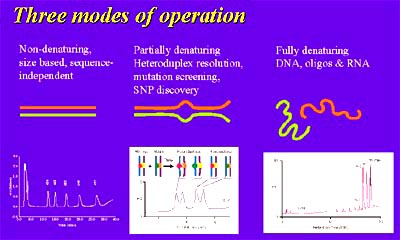
HPLC performs three operation modes.
Under non-denaturing conditions, DNA molecules are fully helical and the differential separation by IP-RP-HPLC is based upon fragment length. Applications that utilize this mode include PCR amplicon analysis, restriction enzyme digest fragments, short tandem repeats (STRs), length polymorphism assays (RFLP, SSLP, etc.), and cloning fragment preparation.
Under partial denaturing conditions (as predicted by the software for the nucleic acid sequence), fragments will separate based on their melting properties. Thus, fragments of the same length that differ in sequence will be resolved. This is useful for discovering and detecting sequence variations (e.g. SNPs, mutations) that may be significant in the predisposition to or development of disease. The method is often called dHPLC or temperature modulated heteroduplex analysis (TMHA) and can result in the high confidence (>95%) detection of a single nucleotide polymorphisms in molecules of up to 1 kilobase.
The third mode of operation, full denaturing, utilizes temperatures at which nucleic acids are single stranded and have no secondary structure. This mode is used to assure quality and purity of oligonucleotides to be used as primers or probes
For more information, call Leslie Linkkila of Transgenomic Inc. at 408-514-3132.
